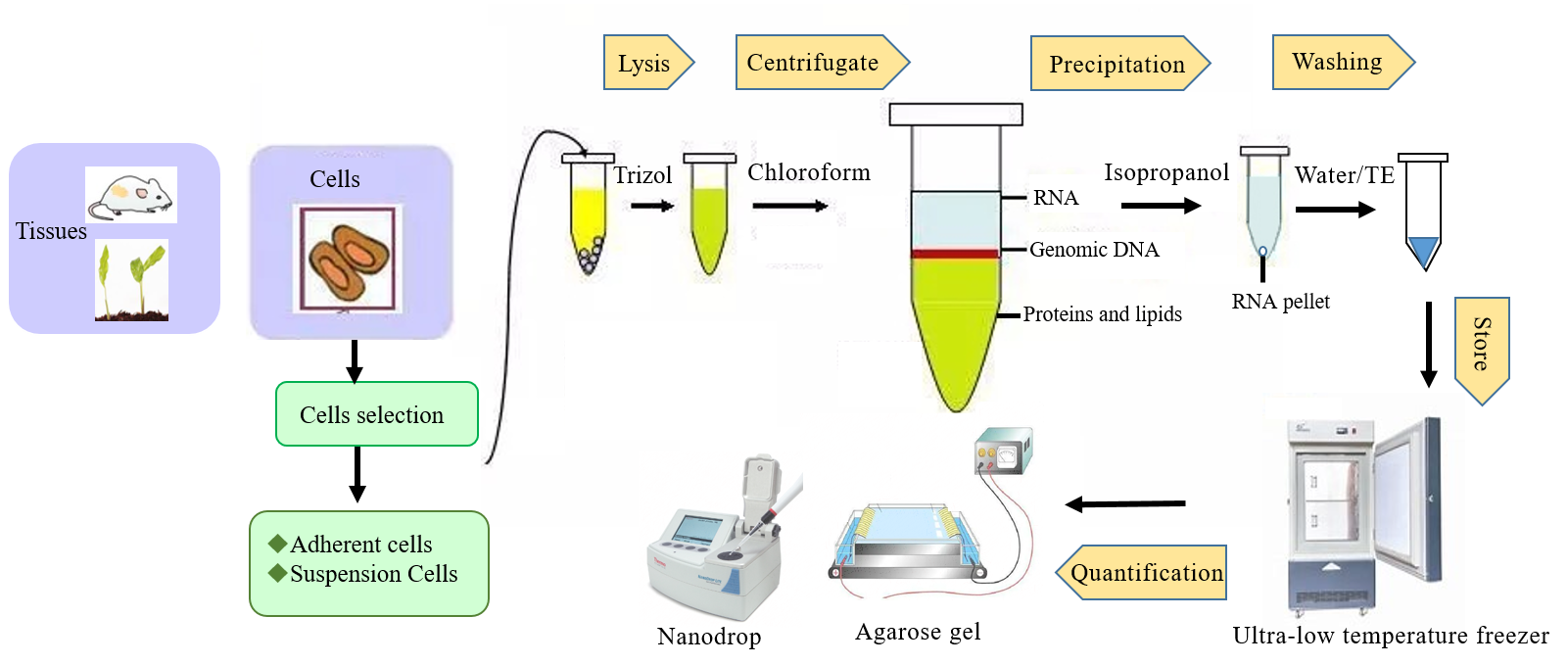
RNA extraction/purification is the isolation of total RNA molecules from biological samples, including cell culturing, tissue, FFPE, blood and total RNA preparations. Acquisition of high-quality RNA is the first and critical step in performing RNA analysis, including PCR, RT-qPCR, Dot blot hybridization, RNA sequencing and northern analysis, etc. There are quite a lot of methods to extract the RNA from various cells and RNA isolation by TRIzol reagent, which is the commonly used and effective to extract the RNA.
1. Freeze 2ml Eppendorf microcentrifuge tube with tissue sample at -80°C and then lyse the sample with the TRIzol reagent. Method to lyse may vary with different cell types. A general guideline for the methods of lyse the cells are as follows.
Table 1. A general guideline for the methods of lyse the cells
|
Biological samples |
Amount of the Sample |
Amount of TRIzol reagent |
Methods to Lyse |
|
Tissues |
50-100 mg |
1 mL |
Grind the tissues with liquid nitrogen and lyse or homogenize tissues by TRIzol in Eppendorf. |
|
Adherent cells |
10 cm2 cells |
1 mL |
Remove the growth media from cell culture plate and add TRIzol directly in it without digest. |
|
Suspension Cells |
1×107 cells |
1 mL |
Resuspend the cells and add TRIzol in it, pipetting up and down for several times. |
2. After adding Trizol to the cells or tissues, place them at room temperature for 5 minutes to allow for dissociation of the nucleoprotein complexes. This step is crucial for the effectiveness of your RNA isolation.
Note: It can be kept at -80℃ for a long time at this stage.
3. Add 0.2 ml chloroform (Chloroform is an organic solvent, which effectively separates the organic phase and the inorganic phase quickly.) per 1 mL TRIzol to the microcentrifuge tube for homogenization. Then invert with your fingers several times and incubate the samples at room temperature for 3 minutes. Centrifugate the sample at maximum speed at 4°C for 15 minutes and three different layers are visible.
Table 2. Three layers containing different contents
|
Upper Aqueous Layer |
RNA |
|
Interphase |
DNA |
|
Bottom Organic Phase |
Proteins and lipids |
Note: Do not use the vortex oscillator, which can lead to the fragmentation of genomic DNA.
4. Take the upper aqueous layer and transfer it to another new Eppendorf by using 200 μl pipette.
Note: Make sure that your pipette tip must not touch the interphase layer. Which is important for extracting high quality RNA.
●RNA precipitation
1. Add 0.5 mL isopropanol per 1 mL TRIzol into the Eppendorf and mix well. Then centrifuge at maximum speed at 4°C for 15 minutes and discard the supernatant. At this time an RNA pellet on the bottom of the Eppendorf will be visible.
●Washing RNA Pellet
1. Add 1ml 75% ethanol per 1 ml Trizol to the RNA pellet, gently shake the centrifuge tube to suspend the precipitate.
2. Centrifuge at 8,000g for 5 minutes at 4°C, then discard the supernatant as much as possible.
3. Let the RNA pellet air dry by keeping the Eppendorf’s lid open for 5-10 min.
Note: Avoid over drying the pellet, otherwise it will be difficult for RNA to dissolve.
●Store the RNA
Immediately add 50 μl RNase free water or TE buffer to resuspend air dried RNA pellet. Pipette it up and down for several times to completely solubilize the RNA and store it at -80℃.
Note: It is suggested that you can make cDNA from this extracted RNA and store cDNA as it is more stable.
●Quantification of RNA
Electrophoresis can be used for the quality check of RNA, including amount and integrity. For more information about the experimental procedure of electrophoresis, please refer to
Protocol for Agarose Gel Electrophoresis. In addition, spectrophotometer also can quantify the RNA. For specific experimental operations, please refer to the instrument manual.
Important tips:
1. Pay attention to wearing masks and gloves all the time to avoid RNA degradation by a large amount of RNases on the surface of our skin. Change new gloves as frequently as possible to make sure the effectiveness of your RNA isolation.
2. Keep work area, equipment, and reagents RNase-free during the whole experiment.
3. RNase-free water should be used to prepare the solution. Add the water and
DEPC into a sterilized glass bottle to a final concentration of 0.1% (v/v), place them overnight and autoclave. Please note that DEPC is carcinogenic and requires careful handling.
If you have any questions,please email us at [email protected].